| Simon: Posters: 5 |

|
Plant & Animal Genome X Meeting - January 12-16, 2002 - http://www.intl-pag.org/
EVALUATION OF GENETIC DIVERSITY AMONG GARLIC CLONES USING MOLECULAR MARKERS: COMPARISON OF AFLPS, RAPDS AND ISOZYMES
Meryem Ipek1 & Philipp Simon1,2
1.Department of Horticulture, University of Wisconsin-Madison, 1575 Linden Dr., Madison, WI , 53706.
2.USDA-ARS Vegetable Crops Research Unit, 1575 Linden Dr. Madison, WI 53706 .
E-mail: psimon@wisc.edu
ABSTRACT
Garlic (Allium sativum L.) is an asexually propagated crop that displays great morphological diversity. We have utilized AFLP and RAPD marker systems to assess the genetic diversity among 45 garlic and three A. longicuspis clones, the proposed progenitor of A. sativum. A subset of 39 clones was also analyzed with four isozyme systems. Three AFLP primer combinations (EcoR I / Mse I), 17 decamer RAPD primers, and four isozyme systems generated a total of unambiguous 183, 80, and eight polymorphic markers, respectively. Jaccard's similarity matrices were prepared for each marker system and used for UPGMA cluster analyses. In agreement with the results of other investigators, A. longicuspis and A. sativum clones were clustered together with no clear separation, suggesting these species are not genetically distinct. Instances of duplicate clones were also identified. The Mantel test indicated a correlation value of 0.96 between AFLP and RAPD, 0.57 between RAPD and isozyme, and 0.55 between AFLP and isozyme. AFLPs are abundant in garlic, they demonstrated much genetic diversity, and they were able to discriminate closely related clones. AFLP is therefore be another tool for detailed assessment of genetic relationships among garlic clones along with RAPD and isozyme markers.
INTRODUCTION
Garlic has been cultivated for at least 5000 years (Hahn 1996) presumably having originated in Central Asia (Vavilov 1951). Although garlic is an asexually propagated crop, it displays great morphological diversity in bulb and leaf size, color and shape, scape presence and height, and flower color, fertility, and bulbil (topset) development in inflorescence (Pooler and Simon 1993).
Until recently, characterization of garlic germplasm has been based primarily on morphological data. However, morphological characters may differ under varying environmental conditions. This can complicate characterization of garlic clones (Bradley et al. 1996; Al-Zahim et al. 1997). To eliminate some of these complications, isozyme and RAPD markers have been utilized to reveal genetic diversity in garlic germplasm (Pooler and Simon 1993; Maass and Klaas 1995; Bradley et al. 1996; Al-Zahim et al. 1997; Lallemand et al. 1997). To characterize the diversity of garlic with greater efficiency, other molecular fingerprinting methods should be utilized. One particularly useful tool to achieve this goal is AFLP which has been used for assessing genetic relationships among garlic germplasm.
MATERIALS AND METHODS
DNA was extracted from freeze-dried, young and healthy leaf samples according to previously described CTAB method (Murray and Thompson 1980).
AFLP protocols were followed according to the manufacturer's procedure with minor modification using theAFLP Kit number I (Gibco-BRL). Half of the recommended reaction volumes were used except pre-selective amplification that was performed at 1/5 the recommended volume. RAPD protocols were performed accordingto the method as described by Bradley et al. (1996), with minor modification. All unambiguously polymorphic bands were identified and scored as present or absent. Similarity matrices were generated according to the coefficient of Jaccard (Sneath and Sokal, 1973) and used to perform UPGMA cluster analysis (Sokal and Michener, 1958) with the software NTSYS-PC version 1.80 (Rohlf, 1993).
Isozyme analysis was carried out according to the methods described by Pooler and Simon (1993). Bands were scored directly from the PAGE gels as present or absent. A similarity matrix prepared using simple matching (SM) coefficient was utilized for UPGMA cluster analysis.
Cophenetic values of dendrograms of AFLP, RAPD, and isozyme were utilized to compare dendrograms by using the Mantel test of significance (Mantel, 1967)
RESULTS
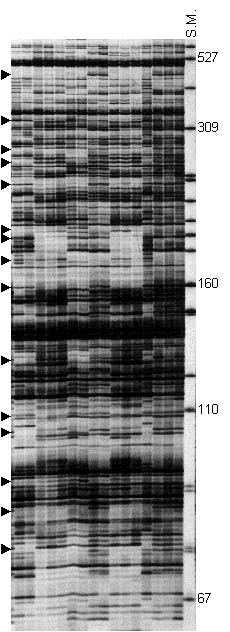
The profile of AFLP fingerprinting patterns of garlic clones with the primer combination I. S. M. is pBR322-Msp I size marker. Arrows point to the bands scored as polymorphic AFLP fragments.
AFLP analysis with three primer combinations generated 64, 63 and 56 unambiguous polymorphic AFLP bands respectively for a total number of 183 polymorphic bands scored with a size range of 67 bp to 457 bp. The Similarity matrix, based on shared fragments of 183 AFLPs, demonstrated a similarity from 0.20 to 1.00 among garlic clones analyzed
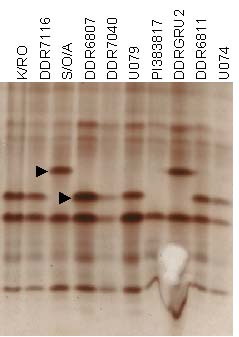
Isozyme pattern of Esterase in 10 garlic accessions. Arrows point to segregating alleles of Esterase.
Eight consistently resolving segregating alleles were scored as present or absent, 3 for G6PDH, 2 each for EST and PGM, and 1 for DIA. SKDH, MDH, IDH, and AAT did not display either consistent or any variation among clones analyzed. Previous studies also indicated that polymorphism in isozyme systems was similarly low (Pooler and Simon 1993; Maass and Klaas 1995).
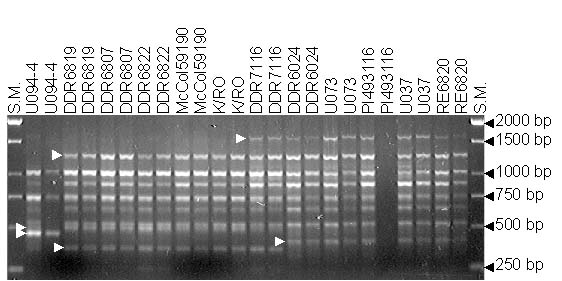
RAPD profile among 12 garlic accession with OPAB-18 primer. Arrows point to polymorphic RAPDs scored on this picture. S.M. is the size markers.
Seventeen polymorphic RAPD primers generated a total of 80 unambiguous polymorphic RAPDs, averaging 4.7 bands per primer. The number of polymorphic amplicons among RAPD primers tested ranged from 2 to 9.
The Dendrogram of 48 clones constructed by UPGMA cluster analysis based on genetic similarity coefficients of shared AFLPs. Roman numericals indicate arbitrary groups.
The dendrogram of UPGMA demonstrated 10 arbitrary groups by clustering clones with more than 60% similarity. Comparing fingerprints of the 48 clones in this study, sixteen clones in six different clusters shared 100% of 183 AFLPs within each group. These clones are likely duplicates.
Groups II, III and IV included non-bolting and incomplete bolting garlic types in which infrequent flowering among the plants from same clone with weak, short, and early senescing flower stalk can be observed. The garlic clones in the rest of groups were bolting garlic types with regular flowering morphology.
A. longicuspis clones, U079 and U094-4, were clustered in group VIII and PI 576914 was clustered in group X with flowering type A. stivum clones. Therefore, our AFLP results suggest that A. longicuspis and A. sativum are not genetically distinct.
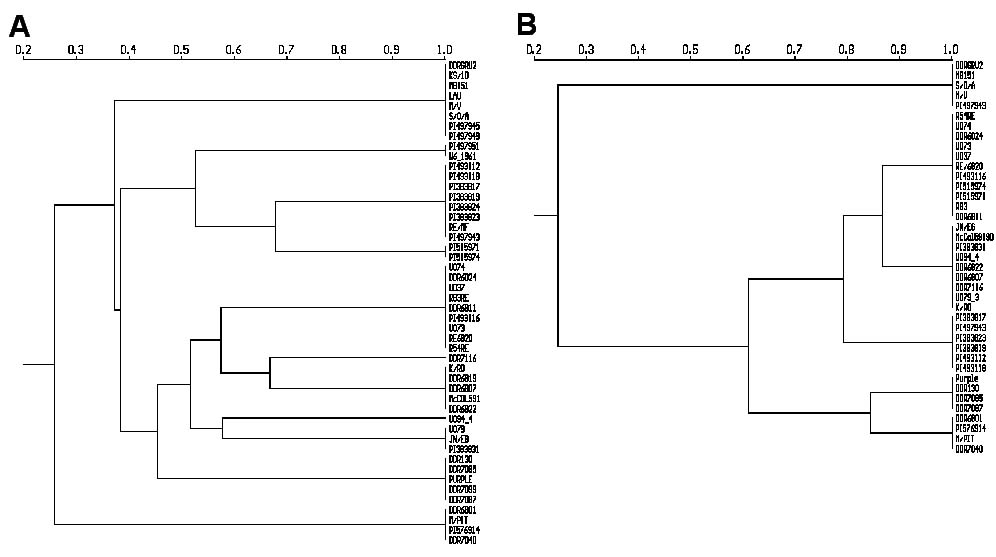
The Dendrograms of UPGMA cluster analysis revealed by A) RAPDs and B) Isozymes
Clones having more than 70% similarity by AFLPs shared 100% of their RAPDs. Above 50% similarity, grouping did not change between dendrograms of AFLP and RAPD. Comparison of AFLP and RAPD dendrograms with Mantel test indicated high concordance between AFLP and RAPD grouping with correlation of 0.96.
Clones sharing more than 80% of the AFLPs were clustered together in the same group by isozyme markers. Cluster analysis of isozyme data generated six groups out of nine arbitrary groups of AFLP and RAPD by combining groups IV and V into one group and groups VI, VII, and VIII into another group. The Mantel test illustrated that correlation between dendrograms of AFLP (39 taxa) and isozyme was 0.56, and similarly correlation between dendrogram of RAPD and isozyme was 0.55 and these correlations indicates significant association.
CONCLUSIONS
![]() AFLPs are numerious in galic and only three primer combinations generated 180 polyporhic AFLPs
AFLPs are numerious in galic and only three primer combinations generated 180 polyporhic AFLPs
![]() Genetic diversity revealed by AFLP analysis was high between groups although putative duplication and tight clustering was observed within groups.
Genetic diversity revealed by AFLP analysis was high between groups although putative duplication and tight clustering was observed within groups.
![]() The UPGMA cluster analysis of AFLP were associated with the flowering phenotype.
The UPGMA cluster analysis of AFLP were associated with the flowering phenotype.
![]() AFLP, RAPD, and isozyme data suggest that A. longicuspis is not genetically distint from A. sativum.
AFLP, RAPD, and isozyme data suggest that A. longicuspis is not genetically distint from A. sativum.
![]() The UPGMA cluster analyses of AFLP, RAPD, and isozyme data were comparable. Correlation between AFLP and RAPD was higher that correlation between AFLP and Isozyme, and RAPD and isozyme, but all correlations were significant.
The UPGMA cluster analyses of AFLP, RAPD, and isozyme data were comparable. Correlation between AFLP and RAPD was higher that correlation between AFLP and Isozyme, and RAPD and isozyme, but all correlations were significant.
![]() AFLP detected polymorphism with highest efficiency although all marker types were informative in this study.
AFLP detected polymorphism with highest efficiency although all marker types were informative in this study.
![]() With abundant AFLPs, a better approximation to the true genetic variation can be detected along with RAPD and Isozyme.
With abundant AFLPs, a better approximation to the true genetic variation can be detected along with RAPD and Isozyme.
References
Al-Zahim, M., H. J. Newbury, and B. V. Ford-Lloyd. 1997. Classification of genetic variation in garlic (Alliumsativum L.) revealed by RAPD. HortScience 32:1102-1104.
Bradley, K. F., M. A. Rieger, and G. G. Collins. 1996. Classification of Australian garlic cultivars by DNA fingerprinting. Aust. J. Exp. Agric. 36:613-618.
Hahn, G. 1996. History, folk medicine and legendary uses of garlic, p. 1-34. In: H. P. Koch and L. D. Lawson (eds.) Garlic, the science and therapeutic application of Alliumsativum L. and related species (2nd edition), Williams and Wilkins, Baltimore, Maryland.
Lallemand, J., C. M. Messian, F. Briad, and T. Etoh. 1997. Delimitation of varietal groups in garlic (Alliumsativum L.) by morphological, physiological and biochemical characters, p. 123-132. In: J. L. Burba and C. R. Galmarini (eds.) Proc. I Int. Symp. Edible Alliaceae, Acta Hort. 433.
Maass, H. I. and M. Klaas. 1995. Infraspecific differentiation of garlic (Alliumsativum L.) by isozyme and RAPD markers. Theor. Appl. Genet. 91:89-97.
Mantel, N.A. 1967. The detection of disease clustering and a generalized regression approach. Cancer Res. 27: 209-220
Murray, J. M. and W. Thompson. 1980. Rapid isolation of high-molecular weight plant DNA. Nucleic Acids Res. 8:4321-4325.
Pooler, M. R. and P. W. Simon. 1993. Characterization and classification of isozyme and morphological variation in a diverse collection of garlic clones. Euphytica 68:121-130.
Rohlf, F. J. 1993. NTSYS-pc numerical taxanomy and multivariate analysis system. Version 1.8. Exeter Publ., Setauket, NY.
Sneath, P. H. A. and R. R. Sokal. 1973. Numerical taxonomy. Freeman, San Francisco.
Sokal, R. R. and C. D. Michener. 1958. A statistical method for evaluating systematic relationships. Univ. Kansas Sci. Bull. 38:1409-1438.
Vavilov, N. I. 1951. The origin, variation, immunity and breeding of cultivated plants. Chronica Botanica 13:1-364.